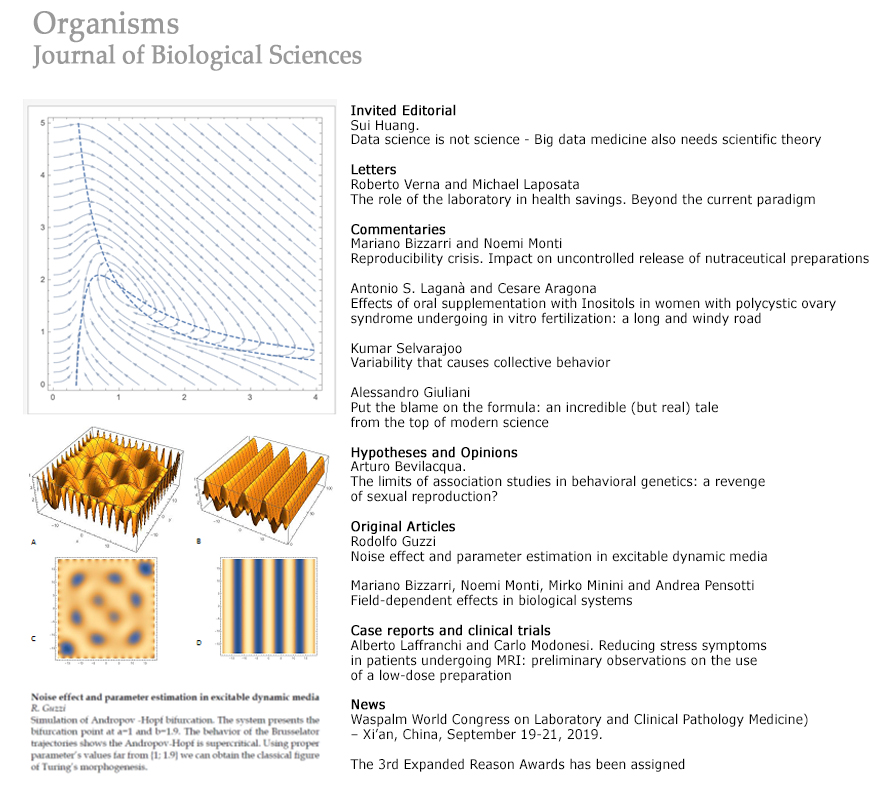
Noise effect and parameter estimation in excitable dynamic media
DOI:
https://doi.org/10.13133/2532-5876_5.7Abstract
Stochasticity in gene expression arises from fluctuations in transcription and translation. This phenomenon has implications for cellular regulation. The novel techniques for single-cell analysis have provided new experimental and theoretical investigations. As a result, a coherent picture of stochasticity in prokaryotic and eukaryotic gene expression has been obtained. In this paper, we analyze the behavior of a stochastic process applied to Brusselator, investigating the noise affecting this system. Once the noise has been retrieved, the maximum likelihood estimation may be used to retrieve the parameters of the system itself. Although these techniques have been applied to simple reaction processes as those hypothesized with Brusselator, the methodology is general and can be used to any reaction and related linear and nonlinear ODEs.Downloads
Published
How to Cite
Issue
Section
License
Copyright Agreement with Authors
Before publication, after the acceptance of the manuscript, authors have to sign a Publication Agreement with Organisms. The authors retain all rights to the original work without any restrictions.
License for Published Contents

You are free to copy, distribute and transmit the work, and to adapt the work. You must attribute the work in the manner specified by the author or licensor (but not in any way that suggests that they endorse you or your use of the work).